Unsupervised Machine Learning allows to explore further the whole transcriptome analysis of human pluripotent stem cells cardiac differentiation
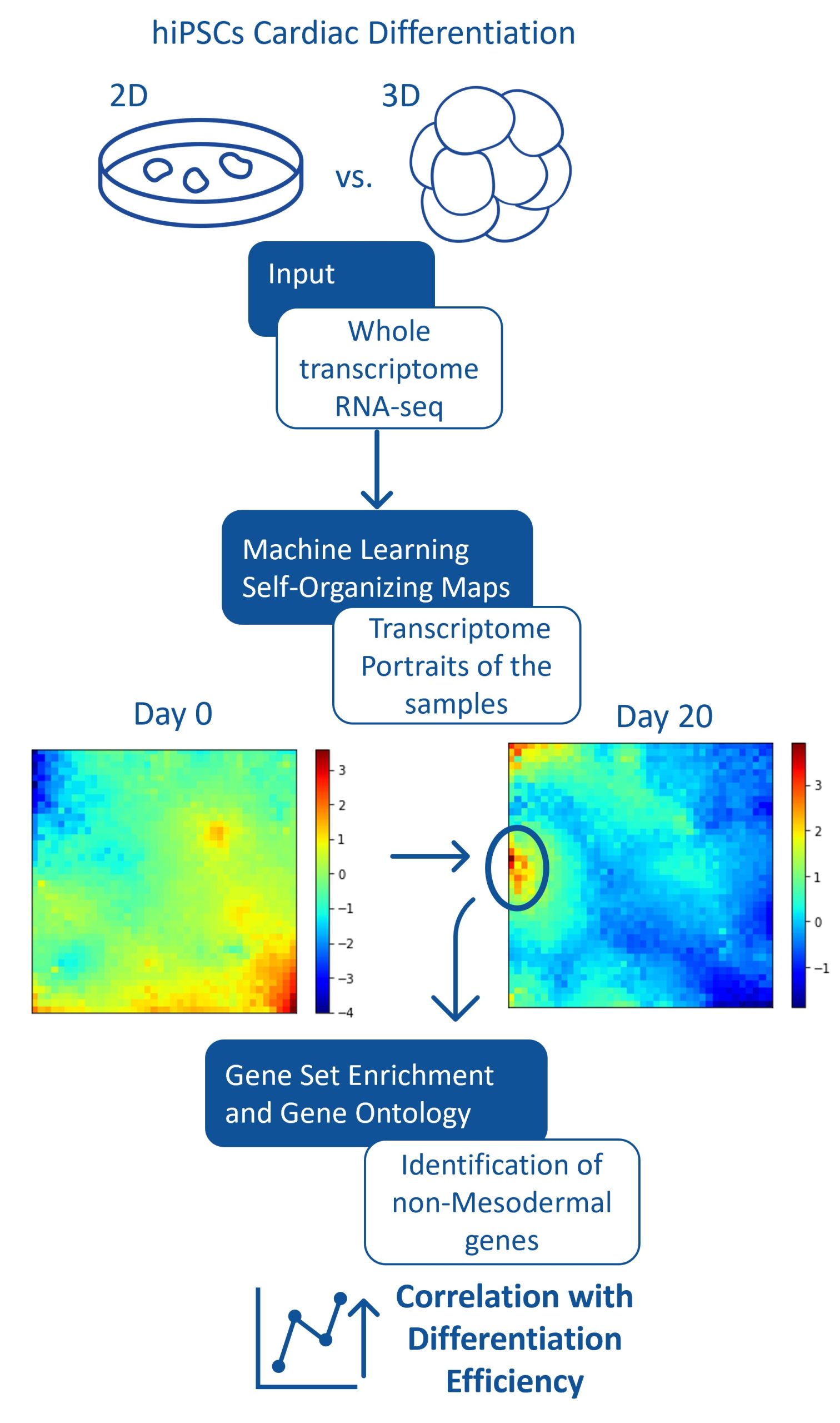
Is it possible to visualize the entire #Transcriptome of hiPSCs undergoing Cardiac differentiation? Can we visually inspect the differences, at gene expression level, between a standard 2D protocol and a novel 3D differentiation strategy? Is the inter-replicate variability worth looking at?
The answer to all these questions is : Yes!
We are delighted to share that you can find all that in our latest publication entitled "Unsupervised analysis of whole transcriptome data from human pluripotent stem cells cardiac differentiation"!
In this study, we applied several unsupervised machine learning algorithms to analyze transcriptomic data from human pluripotent stem cells undergoing cardiac differentiation.
This approach effectively allowed us to portray the changes in gene expression that occurred throughout the differentiation processes, with a visual representation of the entire transcriptome. With this approach, we observed a correlation between low cardiomyocyte differentiation efficiencies and the early expression of a set of non-mesodermal genes.
We expect that this workflow may also be used for studying and understanding other #StemCell differentiation processes. See more.